In our last article, we looked at how we can use Strings in Rust to store Genome data (DNA sequences). We will be using Strings extensively in our future code, as they are very convenient when it comes to storing and manipulating DNA/RNA data.
In this article, we are taking a look at one more, very important data structure that we are going to need for more advanced algorithms. After we compare both Dictionaries (Python) and Hash Maps (Rust), we will implement a DNA reverse_complement function.
Part 1: Dictionary and a Hash Map
Let’s create two new files called data_structures in each folder (Python/Rust), and add an empty functions:
For Python, we create a function dicts() and for Rust, we have _hash_maps() as that is what those data structures are called.
Let’s start by taking a quick look at Python Dictionary, so we have a reference point for our Rust Hash Map version.
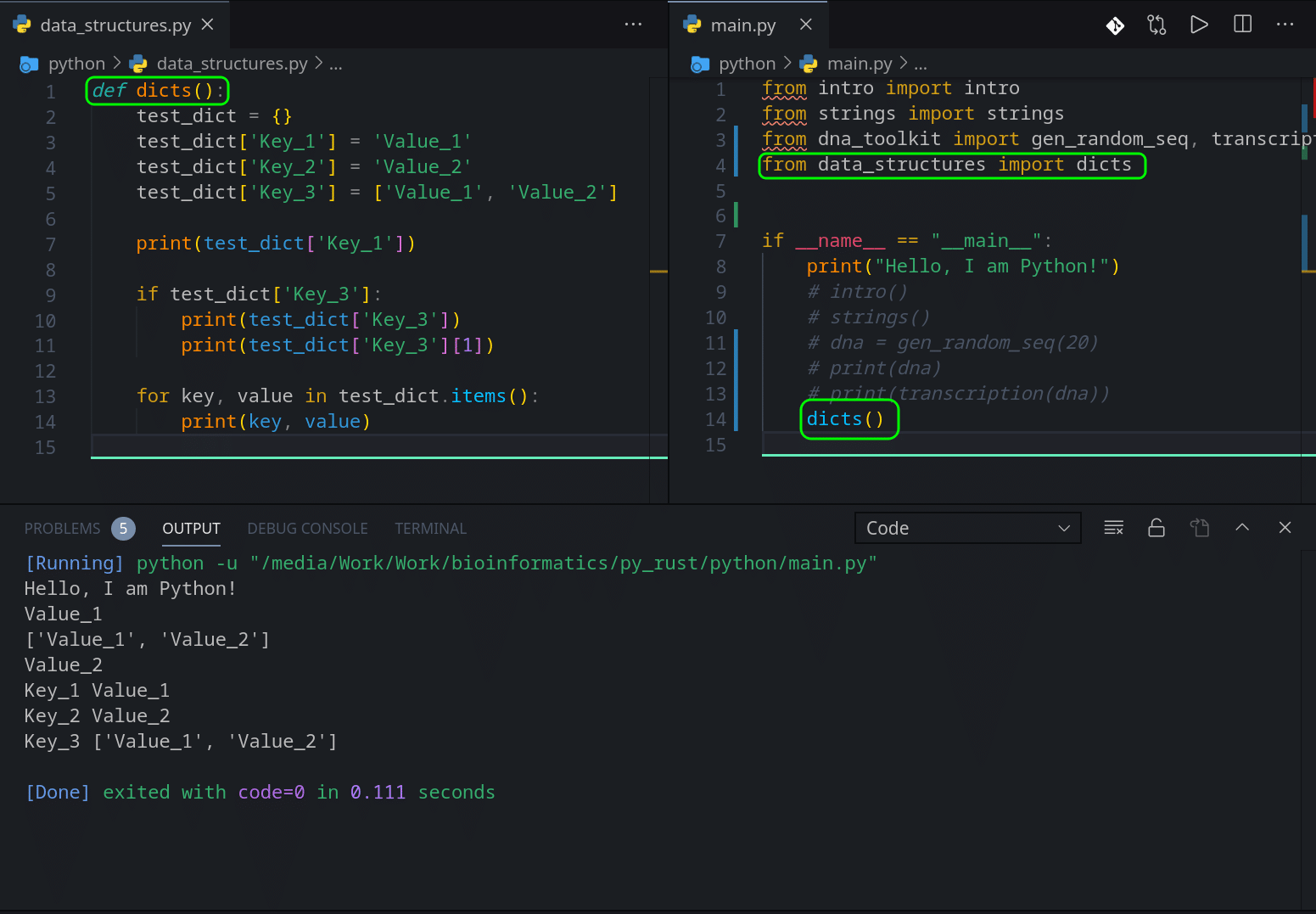
def dicts():
test_dict = {}
test_dict['Key_1'] = 'Value_1'
test_dict['Key_2'] = 'Value_2'
test_dict['Key_3'] = ['Value_1', 'Value_2']
print(test_dict['Key_1'])
if test_dict['Key_3']:
print(test_dict['Key_3'])
print(test_dict['Key_3'][1])
for key, value in test_dict.items():
print(key, value)This is very easy, right? On the line 2 we create an empty dictionary and on lines 3-5 we add Key-Value pairs. Line 5 adds a List as a Value.
On the line 7 we print the Value that ‘Key_1’ is pointing at, which is a string ‘Value_1’.
On the line 9, we make sure that our ‘test_dict’ has a key ‘Key_3’, and if yes, we print its Value. In this case, Line 10 will print a List and the line 11 will print the second value of the list ‘Value_2’.
And the last part is a For loop which accesses ‘items’ of our dictionary, which are Key-Value pairs, iterates through them and prints out both.
If we now call this function in our main.py file, we should see this output:
Now let’s see how we can replicate this in Rust:
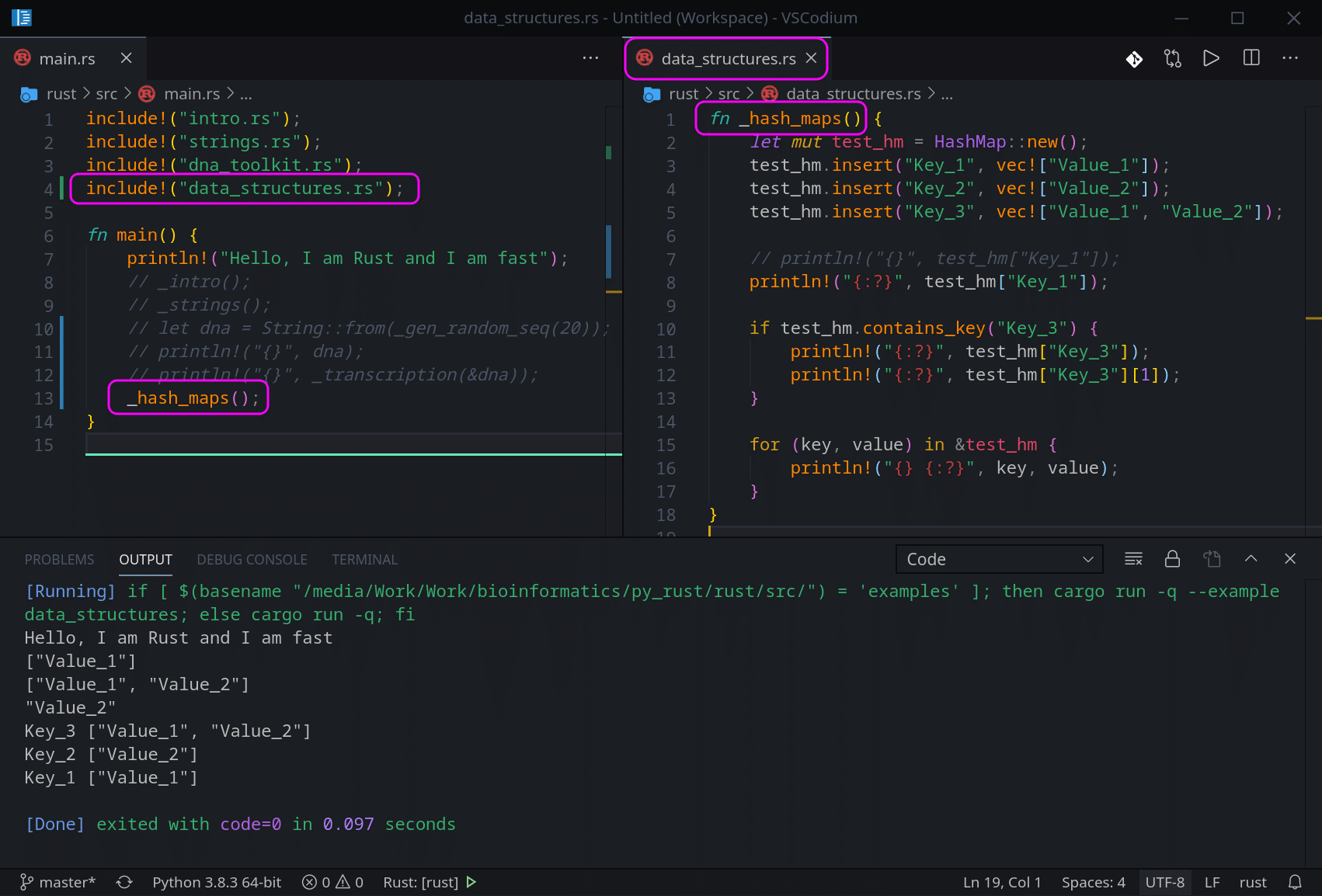
use std::collections::HashMap;
fn _hash_maps() {
let mut test_hm = HashMap::new();
test_hm.insert("Key_1", vec!["Value_1"]);
test_hm.insert("Key_2", vec!["Value_2"]);
test_hm.insert("Key_3", vec!["Value_1", "Value_2"]);
// println!("{}", test_hm["Key_1"]);
println!("{:?}", test_hm["Key_1"]);
if test_hm.contains_key("Key_3") {
println!("{:?}", test_hm["Key_3"]);
println!("{:?}", test_hm["Key_3"][1]);
}
for (key, value) in &test_hm {
println!("{} {:?}", key, value);
}
}On the line 1 we are including a HashMap module as it is not included in Rust by default, unlike Vectors.
On the line 4 we create an empty Hash Map (a dictionary in Python).
On lines 5–7 we insert 3 Key-Value pairs the same way as in Python, but in Rust we need to call ‘insert’ method on our Hash Map. If we want to add a Vector with multiple values, like we do on the line 7, we need to make a Vector the default Value type for the whole Hash Map. This is even if we add a single Value like we do on lines 5 and 6.
Line 9 is there but is commented out. You can see that we are trying to print the Value that has a Key “Key_1”, using ‘{}’. This will generate the following error:
error[E0277]: `std::vec::Vec<&str>` doesn't implement `std::fmt::Display`
--> src/dna_toolkit.rs:24:20
|
24 | println!("{}", test_hm["Key_1"]);
| ^^^^^^^^^^^^^^^^ `std::vec::Vec<&str>` cannot be formatted with the default formatter
|
= help: the trait `std::fmt::Display` is not implemented for `std::vec::Vec<&str>`
= note: in format strings you may be able to use `{:?}` (or {:#?} for pretty-print) instead
= note: required by `std::fmt::Display::fmt`
= note: this error originates in a macro (in Nightly builds, run with -Z macro-backtrace for more info)Amazing that the Rust compiler not only tells us that something is wrong, but also suggests a fix. Let’s use {:?} to format our output, which is a Vector in this case.
The rest of the code is almost identical to Python. The only difference is in Rust, we use ‘contains_key‘ method to check if a Key is in our Hash Map. Last For loop has the same structure, and does not even require items() method like Python does. Here is the final version:
Part 2: DNA Reverse Complement function
Now that we are armed with Dictionaries/Hash Maps, let’s take our existing DNA Reverse Complement function and port it to Rust. In this function we want to map one type of character to another, and given a string, use that map as a translator to generate a new string. We also want to reverse the result before we return it. Let’s go back to dna_toolkit.py/.rs files and add a new function.
So here is our Python version:
def reverse_complement(dna):
"""
Generating a complement string and returning
reveresed version.
"""
trans_dict = {'A': 'T', 'T': 'A', 'C': 'G', 'G': 'C'}
complement_dna = ""
for nuc in dna:
complement_dna += trans_dict[nuc]
return complement_dna[::-1]Nice, clean, and easy, right? This function accepts a string and returns a string.
We just predefine a ‘trans_dict’, translation dictionary on the line 6, which we use as a mapper. On line 7 we create an empty string we will use to accumulate a new set of values in.
For loop on line 9 just loops through each character in the string of dna we passed to that function and checks them against the dictionary, and appends a mapped character to the ‘complement_dna’ string.
Last line 12 returns a reversed version of ‘complement_dna’.
So if we run our code now, we should see this:
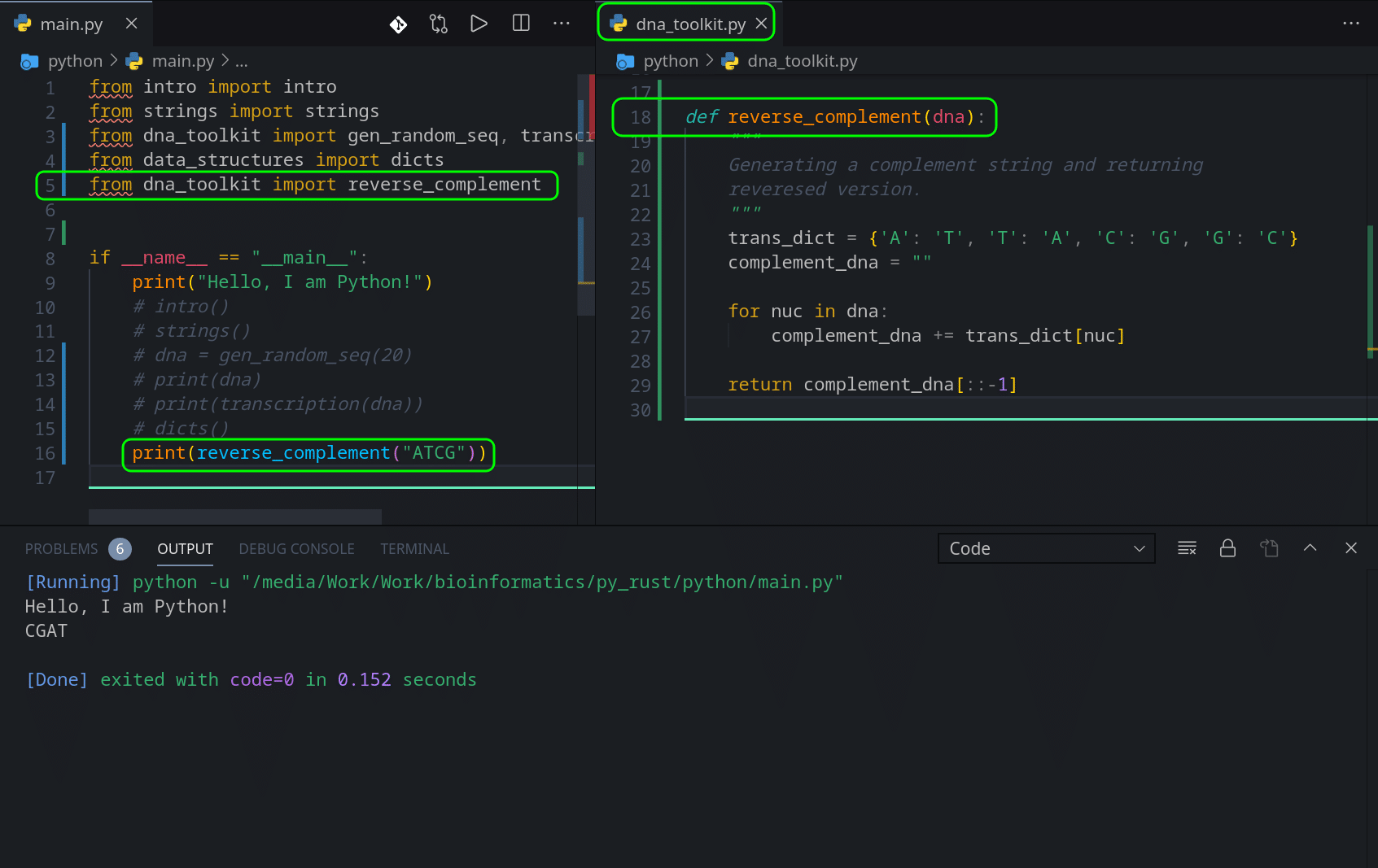
We pass “ATCG” into our new function, it is being mapped against our mapping dictionary and “TAGC” is being generated. A reversed version is returned, and we see ‘CGAT’ in our output window.
Here is ‘Pythonic’ version of the same code. Let’s hope Rust will gain that type of functionality in the future. You could start your own Rust ‘crate’ and replicate this type of functionality and share it with the community.
def reverse_complement(self):
"""
Generating a complement string and returning
reveresed version.
"""
mapping = str.maketrans('ATCG', 'TAGC')
return self.seq.translate(mapping)[::-1]Here is the Rust version:
fn _reverse_complement(dna: &String) -> String {
// Generating a complement string and returning
// reveresed version.
let trans_hashmap: HashMap<char, char> = [('A', 'T'), ('T', 'A'), ('C', 'G'), ('G', 'C')]
.iter()
.copied()
.collect();
let mut complement_dna = String::new();
for nuc in dna.chars().rev() {
complement_dna.push(trans_hashmap[&nuc]);
}
return complement_dna;
}Same as Python, it accepts a string and returns a string. On line 4 we attempt to replicate Python as much as we can. The part after = is an array, so we need to convert it into a collection, copy that collection into a Hash Map and return an iterator. That is why we have those 3 methods after our Hash Map. I suggest you read about these methods here: .iter(), .copied() and here .collect().
Line 9 is a new empty string (from our previous article).
Lines 11, 12 and 15 are almost the same as Python. The only difference being that in Rust, we use ‘dna.chars().rev()‘ to loop through the reversed set of characters from ‘dna‘ and return a string, while In Python we loop through the normal string, and then we reverse it before returning it. Subtle but important difference.
Note, that our mapper ‘trans_hashmap’ is immutable. We can make it mutable and use .insert() method like this:
let mut trans_dict = HashMap::new();
trans_dict.insert('A', 'T');
trans_dict.insert('T', 'A');
trans_dict.insert('C', 'G');
trans_dict.insert('G', 'C');But a static, immutable version should be a faster solution. In this case, we have a fixed set of characters that will not change. So the original approach makes more sense.
Here is the final result:
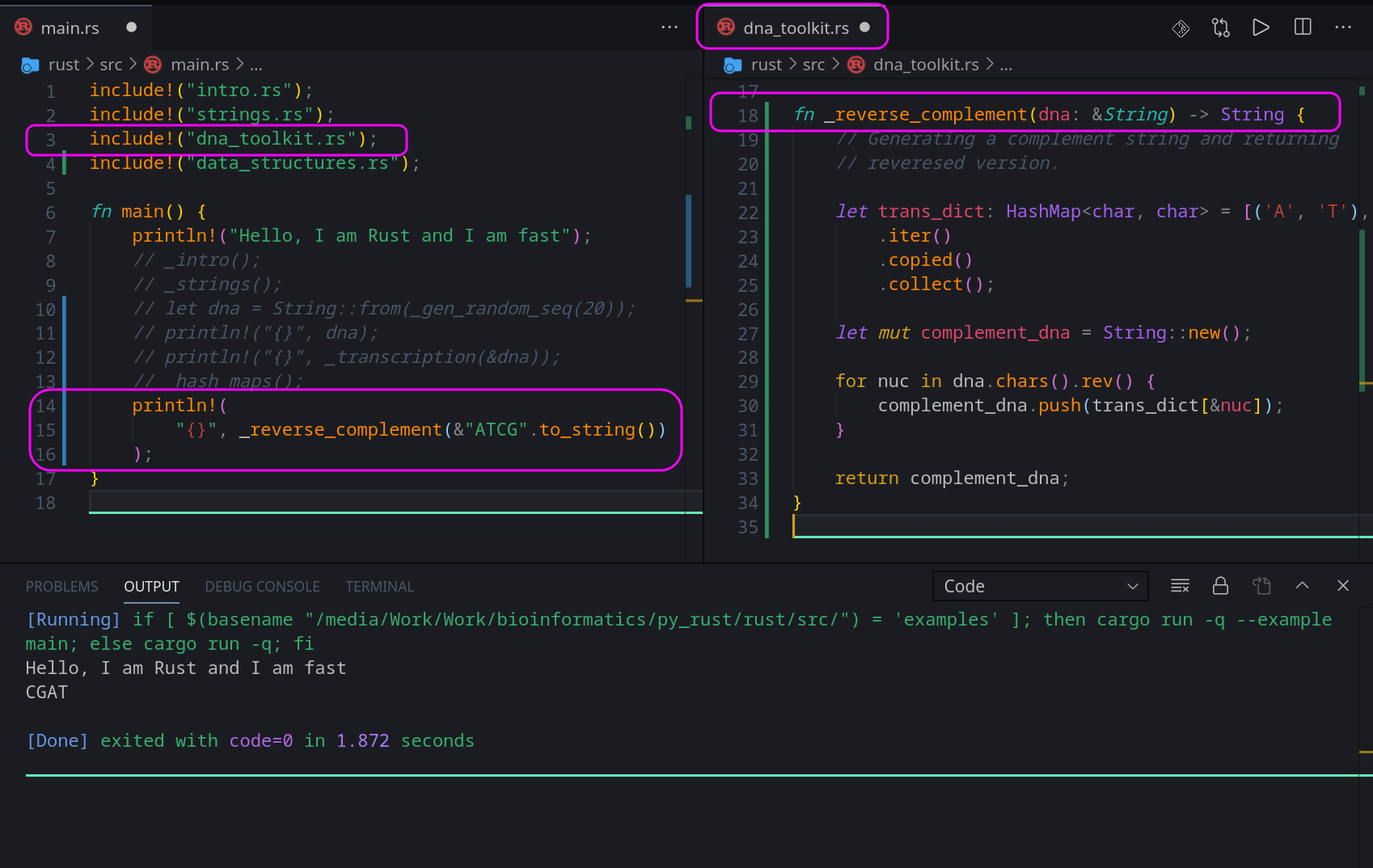
We need to tweak the value we pass to be printed in Rust. If we try to pass just “ATCG”, Rust will tell us that we are passing a slice ‘str’ but our function is expecting a reference to a string ‘&String’. So we need to convert ‘str’ to a String and pass it by reference.
Now let’s finish by using our random DNA generator and look at the final result for both languages:
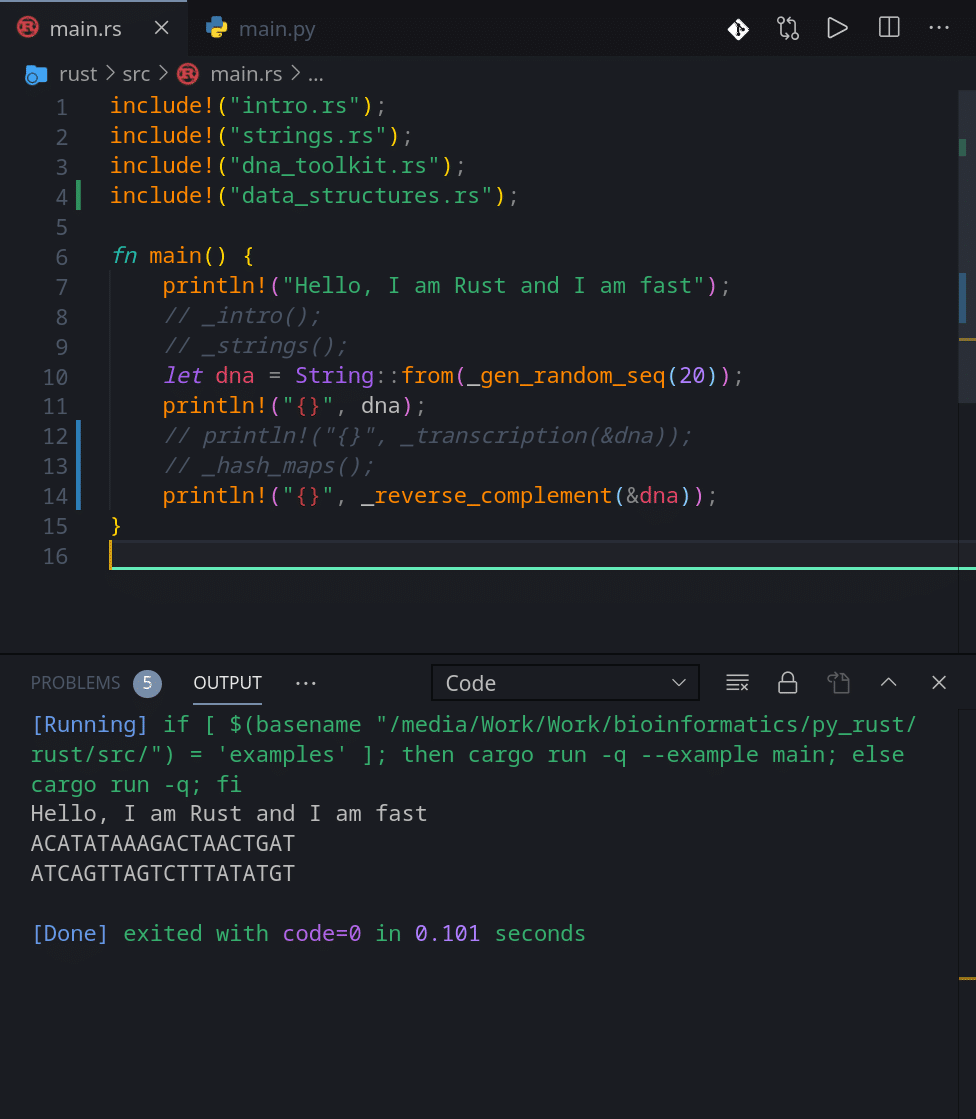
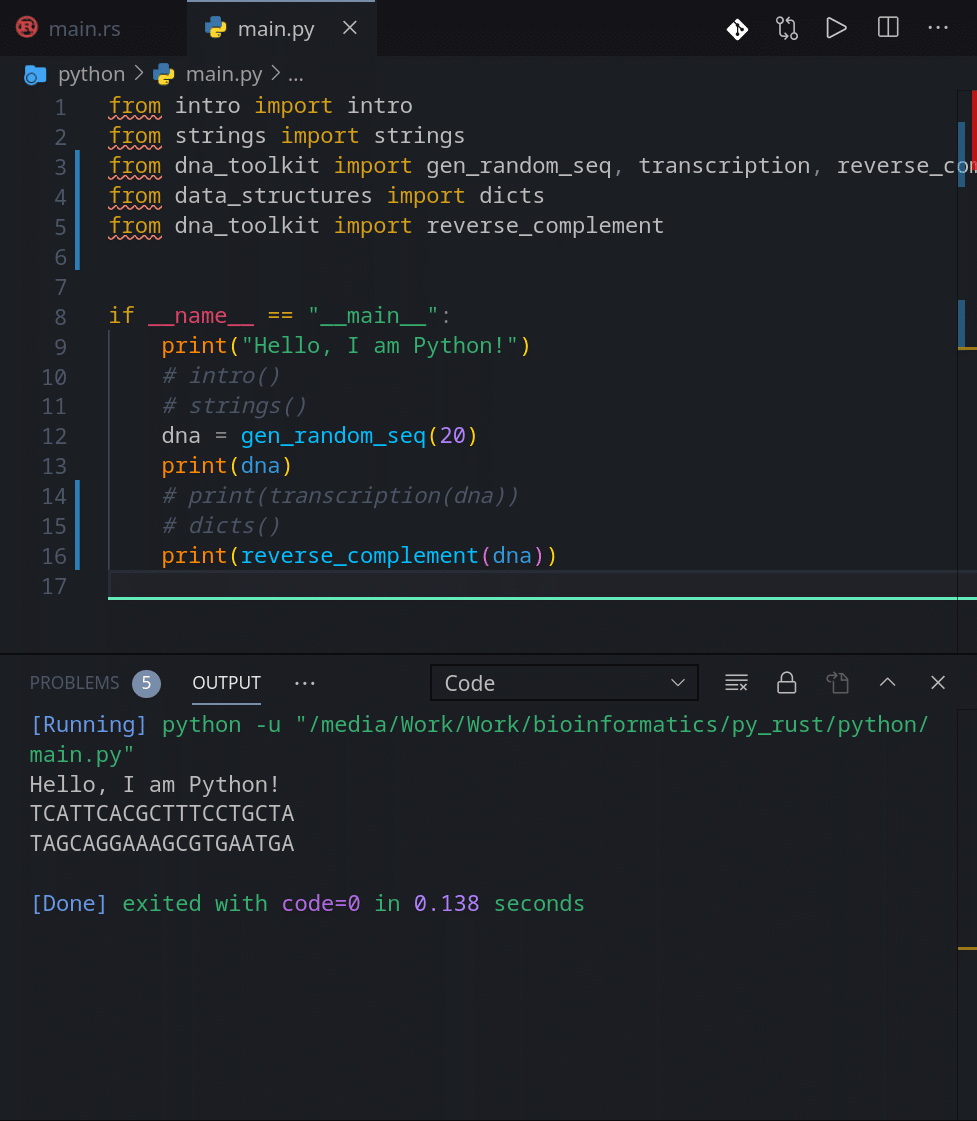
Alright. This is it for this article. See you in the next one.
Links
- Rust HashMap video #1: https://youtu.be/sTK8fagTsMk
- Rust HashMap video #2: https://youtu.be/ic9WEuto-gE
GitLab: https://gitlab.com/RebelCoder/py_rust.git
Recommended Rust programming book:

(UK / US)
For a video versions of these articles, check out my playlist here:
