Welcome back! Today we continue working on our DNA Toolkit project. In our last article, we created the first two functions: validate_seq and nucleotide_frequency. We are not going to change the file structure, though we will add two more functions and one data structure. The functions we add today will finally replicate a real biological process. DNA into RNA Transcription and Complement Generation (Reverse Complement for computational purpose).
DNA is an amazing way for nature to store instructions, it is biological code. It is efficient for computational use in two ways. Firstly, by having just a single strand of DNA we can generate another strand using a complement rule, and secondly, that data is very compressible. We will look into DNA data compression in our future articles/videos.
In the two images below, we can see these three steps:
- DNA Complement generation.
- DNA -> RNA Transcription.
- RNA -> Polypeptide -> Protein Translation.
In this article, we will implement the first two steps, marked 1 and 2 in the second image.


We start by adding one new structure DNA_ReverseComplement to our structures.py file as a Python dictionary. This dictionary will be used when we go through a DNA string, nucleotide by nucleotide. The dictionary will then return a complementary nucleotide. This approach is easy to understand as we just used a dictionary and a for loop. There is a more Pythonic way of generating a complement string, which is discussed later,
DNA_Nucleotides = ['A', 'C', 'G', 'T']
DNA_ReverseComplement = {'A': 'T', 'T': 'A', 'G': 'C', 'C': 'G'}Now, let’s add a function that will use this dictionary to give us a complementary DNA strand and reverse it.
def reverse_complement(seq):
"""
Swapping adenine with thymine and guanine with cytosine.
Reversing newly generated string
"""
return ''.join([DNA_ReverseComplement[nuc] for nuc in seq])[::-1]Here we use a list comprehension to loop through every character in the string, matching it with a Key in DNA_ReverseComplement dictionary to get a Value from that dictionary. When we have a new list of complementary nucleotides generated, we use ‘’.join method to glue all the characters into a string and reverse it with [::-1].
So this approach is easy to read and understand. The code looks more structured as we used our predefined structures file. It is also translatable to other programming languages that way.
Let’s try using Pythonic, maketrans method to solve this problem without even using a dictionary.
def reverse_complement(seq):
"""
Swapping adenine with thymine and guanine with cytosine.
Reversing newly generated string
"""
# Pythonic approach. A little bit faster solution.
mapping = str.maketrans('ATCG', 'TAGC')
return seq.translate(mapping)[::-1]Python string method maketrans() returns a translation table that maps each character in the intabstring into the character at the same position in the outtab string. Then this table is passed to the translate() function.
Note − Both intab and outtab must have the same length.
You can learn more about this method here: Python String maketrans() Method
Next, we define the Transcription function. It is a very simple, one-liner in Python:
def transcription(seq):
"""
DNA -> RNA Transcription.
Replacing Thymine with Uracil
"""
return seq.replace("T", "U")This code is self-explanatory, as we are using standard Python language functionality so far. Here, we just find every occurrence of Thymine and replace it with Uracil. We also return a new RNA string, without effecting the original DNA string, passed through seq.
You might have noticed we started using an interesting code commenting approach. We wrap our comments into:
""" comment """ instead of # comment
This is called a Dockstring. It is super useful when you have to write a complex algorithm, or you have many parameters your function accepts. By adding a description in a form of a Dockstring, the code editor will show you that information when you call that function. For a demonstration of how this works, check out this Video.
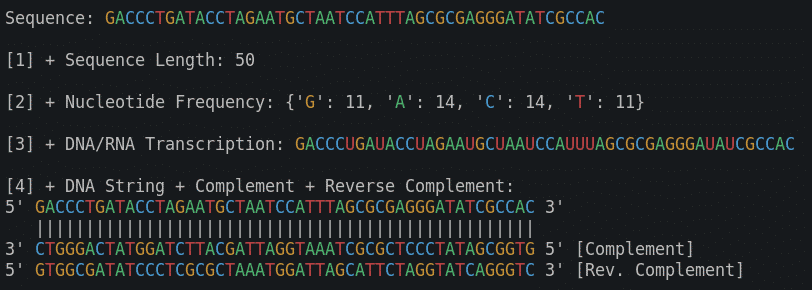
This is it. We are done implementing our two new functions. Let’s test them by adding the output from both to our main.py file. We will use f-strings again to nicely format the output.
print(f'[3] + DNA/RNA Transcription: {transcription(DNAStr)}\n')
print(f"[4] + DNA String + Complement + Reverse Complement:\n5' {DNAStr} 3'")
print(f" {''.join(['|' for c in range(len(DNAStr))])}")
print(f"3' {reverse_complement(DNAStr)[::-1]} 5' [Complement]")
print(f"5' {reverse_complement(DNAStr)} 3' [Rev. Complement]\n")So here is the output for all 4 functions we implemented so far:
Sequence: CCGTTGGATGGGCTAGTTCGTCCTTCCTACACCAGTGCAGGTGCGGTTTA
[1] + Sequence Length: 50
[2] + Nucleotide Frequency: {'C': 13, 'G': 15, 'T': 15, 'A': 7}
[3] + DNA/RNA Transcription: CCGUUGGAUGGGCUAGUUCGUCCUUCCUACACCAGUGCAGGUGCGGUUUA
[4] + DNA String + Complement + Reverse Complement:
5' CCGTTGGATGGGCTAGTTCGTCCTTCCTACACCAGTGCAGGTGCGGTTTA 3'
||||||||||||||||||||||||||||||||||||||||||||||||||
3' GGCAACCTACCCGATCAAGCAGGAAGGATGTGGTCACGTCCACGCCAAAT 5' [Complement]
5' TAAACCGCACCTGCACTGGTGTAGGAAGGACGAACTAGCCCATCCAACGG 3' [Rev. Complement]As a bonus, I have added coloring to our code, into a new file: utilites.py. This is not Bioinformatics related, but if you want to practice your Python, and add a function like that, you can view a video version of DNA Toolkit. Part 2 to see how this is done. We will add many more helper functions to utilites.py in the future. Functions for reading/writing files, reading/writing databases, etc.
Here is a GitLab Link for this article: Link
A video version of this article can be viewed here:
This is it for now. See you in the next article.
One thought on “DNA Toolkit Part 2: Transcription and Reverse Complement”